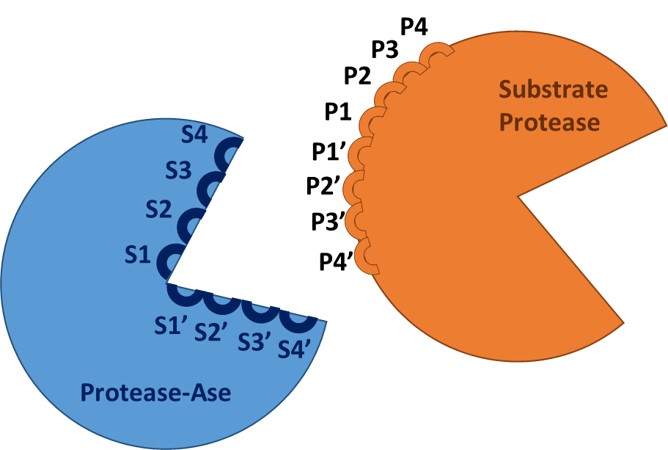
This work has been published by Ferrall-Fairbanks et al in Protein Science.
As an example, find the link to the specificity matrix of cathepsin S: https://www.ebi.ac.uk/merops/cgi-bin/pepsum?id=C01.034;type=P
As an example to test, use the sequence for cathepsin K:
MWGLKVLLLPVVSFALYPEEILDTHWELWKKTHRKQYNNKVDEISRRLIWEKNLKYISIHNLEASLGVHTYELAMN
HLGDMTSEEVVQKMTGLKVPLSHSRSNDTLYIPEWEGRAPDSVDYRKKGYVTPVKNQGQCGSCWAFSSVGALEGQL
KKKTGKLLNLSPQNLVDCVSENDGCGGGYMTNAFQYVQKNRGIDSEDAYPYVGQEESCMYNPTGKAAKCRGYREIP
EGNEKALKRAVARVGPVSVAIDASLTSFQFYSKGVYYDESCNSDNLNHAVLAVGYGIQKGNKHWIIKNSWGENWGN
KGYILMARNKNNACGIANLASFPKM